Grandparent inference from genetic data
Grandparent inference from genetic data: The potential for parentage-based tagging programs to identify offspring of hatchery strays
Thomas A. Delomas and Matthew R. Campbell
Abstract
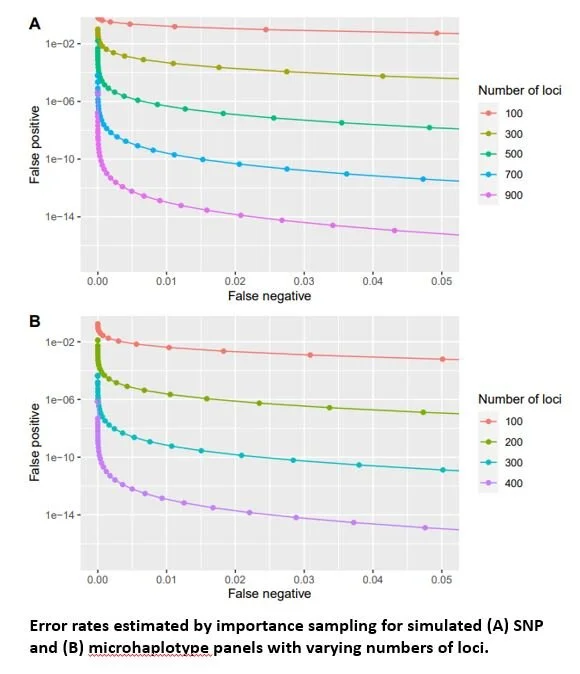
Fisheries managers routinely use hatcheries to increase angling opportunity. Many hatcheries operate as segregated programs where hatchery-origin fish are not intended to spawn with natural-origin conspecifics in order to prevent potential negative effects on the natural-origin population. Currently available techniques to monitor the frequency with which hatchery-origin strays successfully spawn in the wild rely on either genetic differentiation between the hatchery- and natural-origin fish or extensive sampling of fish on the spawning grounds. We present a method to infer grandparent-grandchild trios using only genotypes from two putative grandparents and one putative grandchild. We developed estimators of false positive and false negative error rates and showed that genetic panels containing 500 - 700 single nucleotide polymorphisms or 200 - 300 microhaplotypes are expected to allow application of this technique for monitoring segregated hatchery programs. We discuss the ease with which this technique can be implemented by pre-existing parentage-based tagging programs and provide an R package that applies the method.